Documentation
ASGAL is composed of different modules written in c++ and python. It has been developed and tested on Ubuntu Linux, but should work on any *nix-based system.
Installation
To compile ASGAL and the 3rd party libraries it uses (sdsl-lite and lemon), install:
On an ubuntu system (18.04), the following commands suffice:
sudo apt update
sudo apt install -y build-essential cmake make wget git python3-pip samtools zlib1g-dev python3 python3-pip python3-setuptools python3-biopython python3-biopython-sql python3-pysam python3-pandas python3-setuptools
pip3 install --user gffutils
Then, to download and compile sdsl-lite, lemon, Salmon and ASGAL:
git clone --recursive https://github.com/AlgoLab/galig.git
cd galig
make prerequisites
make
This creates the executable in the bin folder. The python scripts, instead, are in the scripts folder.
Input
ASGAL takes as input:
- a reference chromosome (in FASTA format)
- a gene annotation (in GTF format)
- an RNA-Seq sample (in FASTA or FASTQ format, it can be gzipped)
Note 1 (genome-wide analysis)
ASGAL tool takes as input the annotation of a single gene and the relative chromosome: if you want to use the tool in a genome-wide analysis, please refer to this page.
Note 2 (paired-end sample)
At the moment, ASGAL is not able to directly manage paired-end samples: the only way to use both the fastq files (the reverse and the forward one) consists in merging them into a unique fastq file and then using this file as input for ASGAL. In this way, ASGAL will align each read independently and then it will use the alignments to detect the AS events. Note that this is needed only if you run ASGAL on a single gene: if you use the genome-wide pipeline, the merge is done automatically.
Usage
ASGAL is composed of three different modules, each one performing a different task. We made available a script that runs the full pipeline.
Full Pipeline Script
To run ASGAL pipeline on a single gene, run the following command:
./asgal -g [genome] -a [annotation] -s [sample] -o outputFolder
This command will produce four files in the output folder:
- a .mem file containing the alignments to the splicing graph
- a .sam file, containing the alignments to the splicing graph mapped to the reference genome
- a .events.csv file, containing the alternative splicing events detected in the RNA-Seq sample
- a .log file, containing the log of the execution
We will now specify in more detail the three steps of ASGAL pipeline.
Step 1 - Splice-Aware Aligner
To build the splicing graph of the input gene and to align the input sample to it, run the following command:
./bin/SpliceAwareAligner -g [reference] -a [annotation] -s [sample] -o outputFolder/output.mem
In this way, the alignments to the splicing graph are computed and stored in the output.mem file.
Step 2 (optional) - SAM Formatter
The file obtained in the previous step can be converted into a SAM file (that can be open, for example, with IGV) using:
python3 ./scripts/formatSAM.py -m output.mem -g [reference] -a [anotation] -o outputFolder/output.sam
Observation
The SAM file produce by ASGAL (output.sam) contains the alignments to the splicing graph mapped to the reference genome. These alignments must not be confused with the spliced alignments to the reference genome computed by any spliced aligner (such as STAR or HISAT2). Indeed, our alignments could contain long insertions representing the portions of reads that could align to an intron (and that could represent, for example, a cassette exon).
Step 3 - Events Detector
To analyze the alignments and detect the possible presence of alternative splicing events, run:
python3 ./scripts/detectEvents.py -g [reference] -a [annotation] -m output.mem -o outputFolder/output.events.csv
Output Format
The alternative splicing events are stored in the file output.events.csv, a space-separated value file where each line represents an event. Each alternative splicing event is associated to the intron inducing it and it is described by:
- type of event:
- ES, Exon Skipping
- A3, Alternative acceptor (3’) site
- A5, Alternative donor (5’) site
- IR, Intron Retention
- genomic positions (start and end) of the intron inducing the event
- number of reads confirming the event
- list of annotated transcripts with respect to which the event is expressed
For example,
| Type | Start | End | Support | Transcripts |
|---|---|---|---|---|
| ES | 2000 | 2500 | 150 | Transcript1/Transcript2/Transcript3 |
this entry describes an exon skipping event, induced by 150 reads that support the intron starting at position 2000 and ending at position 2500. The event is expressed with respect to the annotated transcript Transcript1, Transcript2, and Transcript3.
Example
We want align the RNA-Seq sample contained in the example folder to gene CG13375 of Drosophila Melanogaster.
cd example
# Extract the RNA-Seq sample
tar xfz input.tar.gz
# Download reference and annotation from ensembl
wget ftp://ftp.ensembl.org/pub/release-91/fasta/drosophila_melanogaster/dna/Drosophila_melanogaster.BDGP6.dna.chromosome.X.fa.gz
gunzip Drosophila_melanogaster.BDGP6.dna.chromosome.X.fa.gz
mv Drosophila_melanogaster.BDGP6.dna.chromosome.X.fa input/genome.fa
wget ftp://ftp.ensembl.org/pub/release-91/gtf/drosophila_melanogaster/Drosophila_melanogaster.BDGP6.91.chr.gtf.gz
gunzip Drosophila_melanogaster.BDGP6.91.chr.gtf.gz
# Extract the annotation of the gene of interest
grep "CG13375" Drosophila_melanogaster.BDGP6.91.chr.gtf > input/annotation.gtf
rm Drosophila_melanogaster.BDGP6.91.chr.gtf
# Run ASGAL
../asgal -g input/genome.fa -a input/annotation.gtf -s input/sample_1.fa -o CG13375
We should obtain a novel exon skipping events:
Type,Start,End,Support,Transcripts
ES,287042,289040,411,FBtr0300326/FBtr0070103/FBtr0342963
Observation
The correctness of the output can be verified by opening the genome, the annotation and the alignments with IGV and by generating a sashimi plot:
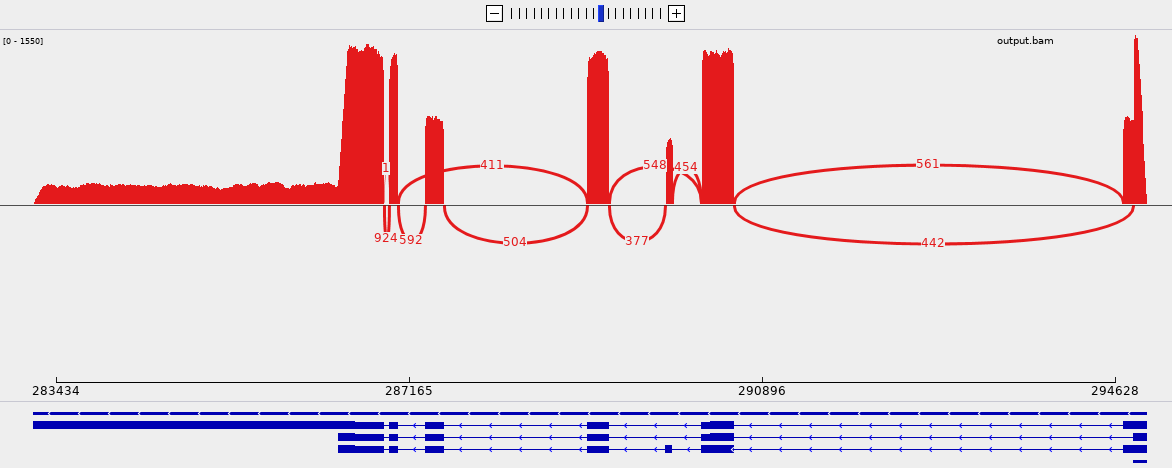
From the picture, we can see that 411 reads support the skipping of the third exon and this event involves the three transcript of the gene.
Command Line Arguments
- all programs show usage information with -h (--help)
Full Pipeline Script
File:
./asgal
Required parameters:
-g,--genome INFILE FASTA input file containing the reference
-a,--annotation INFILE GTF input file containing the gene annotation
-s,--sample INFILE FASTA/Q input file containing the RNA-Seq reads (can be a gzipped file)
-o,--output OUTFOLD output folder
Optional parameters:
-l,--L <int> minimum lenght of MEMs used to build the
alignments (default: 15)
-e,--eps <int> error rate, a value from 0 to 100 used to
compute the maximum number of allowed errors
(default: 3)
-w,--support <int> minimum number of reads needed to confirm an event
(default: 3)
--allevents output all events, not only the novel ones
(default: false, ie only novels)
--verbose verbose print (default: False)
--debug debug print (default: False)
Splice-Aware Aligner
File:
./bin/SpliceAwareAligner
Required parameters:
-g,--genome INFILE FASTA input file containing the reference
-a,--annotation INFILE GTF input file containing the gene annotation
-s,--sample INFILE FASTA/Q input file containing the RNA-Seq reads (can be a gzipped file)
-o,--output OUTFILE output file containing the alignments to the
splicing graph
Optional parameters:
-l,--L <int> minimum lenght of MEMs used to build the
alignments (default: 15)
-e,--eps <int> error rate, a value from 0 to 100 used to
compute the maximum number of allowed errors
(default: 3)
Observations
- an higher value of L improves the speed but reduces the number of aligned reads
- the number of allowed errors is computed for each read as the ratio between the error rate and its length
SAM Formatter
File:
./scripts/formatSAM.py
Required parameters:
-g,--genome INFILE FASTA input file containing the reference
-a,--annotation INFILE GTF input file containing the gene annotation
-m,--mems INFILE input file containing the alignments to
the splicing graph
-o,--output OUTFILE SAM output file
Optional paramter:
-e,--erate <int> error rate, a value from 0 to 100 (default: 3)
Observations
- the error rate should be the same used to compute the alignments
Events Detector
File:
./scripts/detectEvents.py
Required parameters:
-g,--genome INFILE FASTA input file containing the reference
-a,--annotation INFILE GTF input file containing the gene annotation
-m,--mems INFILE input file containing the alignments to
the splicing graph
-o,--output OUTFILE output file containing the events
Optional parameters:
-e,--erate <int> error rate, a value from 0 to 100 (default: 3)
-w,--support <int> minimum number of reads needed to confirm an event
(default: 3)
--allevents output all events, not only the novel ones
(default: only novels)
Observation
- the error rate should be the same used to compute the alignments
- by default, ASGAL outputs only the alternative splicing events that are novel with respect to the given annotation, that are the the events induced by an intron not contained in the annotation